HBV Tool Results
No further algorithm is linked in the HIV-GARDE-tool for the interpretation of HBV sequences. Only on the very bottom of the page you can find a button to directly send the sequence to the geno2pheno-HBV interpretation tool.
In the results view the top section provides information about the sequence name, the sequence date (which corresponds to the date of analysis on the HIV-GRADE website).
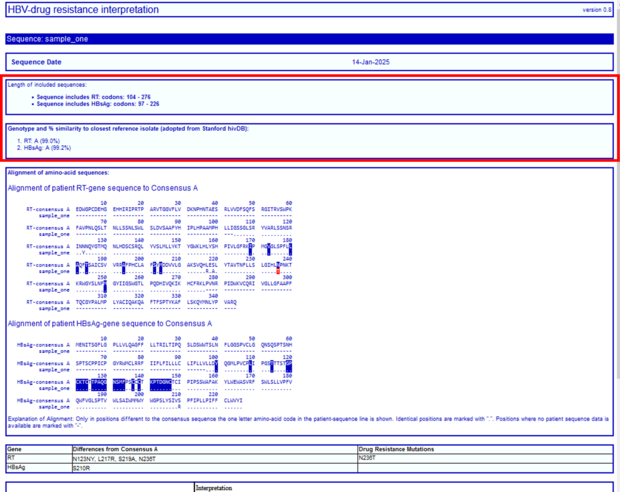
In the next section you will find information about the sequence itself, e.g. to which region the HBV sequence belongs to, which amino acid codons were analyzed and a prediction of the HBV genotype with the percentage of similarity to the closest reference sequence adopted from NCBI sequence data. In our example, the submitted sequence corresponds to HBV RT, covers the amino acid codons 104-264, and HBsAg covers aa 97-226. Sequence is predicted to be HBV genotype A (99% or 99.2%).
If you have selected the “Mutation List Analysis”, the sequence information including the subtype will of course be omitted.
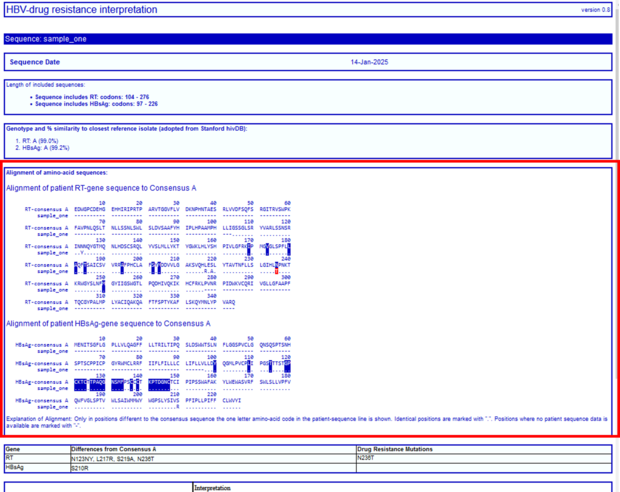
Below the subtype information you will find an alignment of the sequence to the genotype specific wildtype. All resistance relevant mutations are signed in blue, all detected resistance relevant mutations are signed in red.
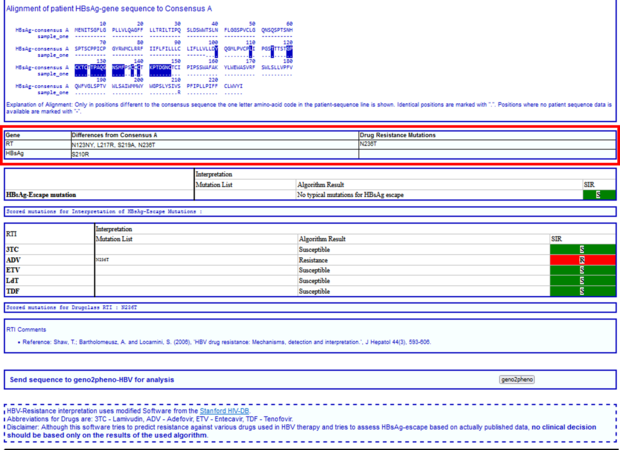
Next, you will find all amino acid differences of your submitted sequence compared to the consensus sequence and on the right hand the “Drug Resistance Mutations”.
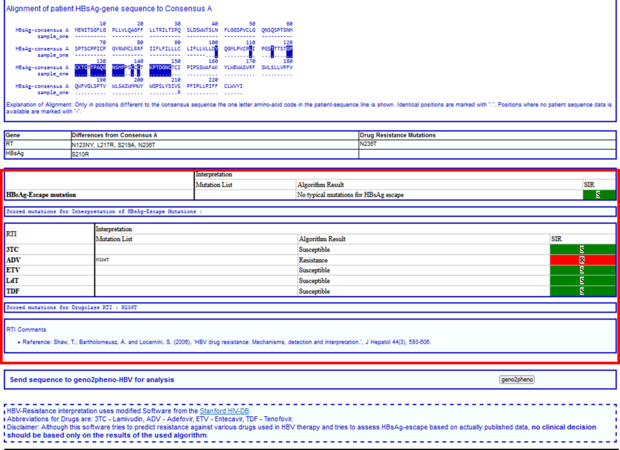
In the next section you will find a table with the sequence interpretation by HIV-GRADE, first the HBsAg-Escape variants, and second the resistance mutations and SIR interpretation for the different HBV drugs.

