HIV Tool Results
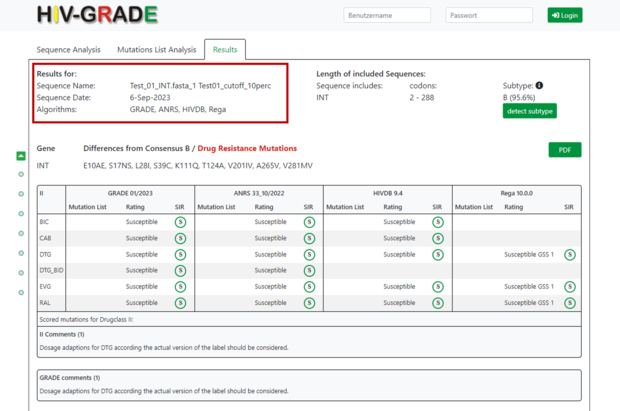
In the results view, for each submitted sequence, the top section provides information about the sequence name, the sequence date (which corresponds to the date of analysis on the HIV-GRADE website), and information about the algorithms used for the analysis.
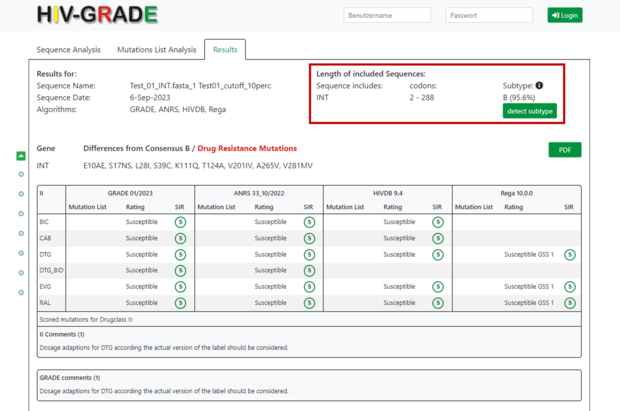
On the right side you will find information about the sequence itself, e.g. to which region of the HIV-1 genome the sequence belongs, which amino acid codons were analyzed and a prediction of the HIV-1 subtype and % of similarity to the closest reference sequence adopted from Standford HIVDB. In our example, the submitted sequence corresponds to HIV-1 integrase (INT), covers the amino acid codons 2-288, and is predicted to be HIV-1 subtype B (95.6%).
If you have selected the “Mutation List Analysis”, the sequence information including the subtype will of course be omitted.
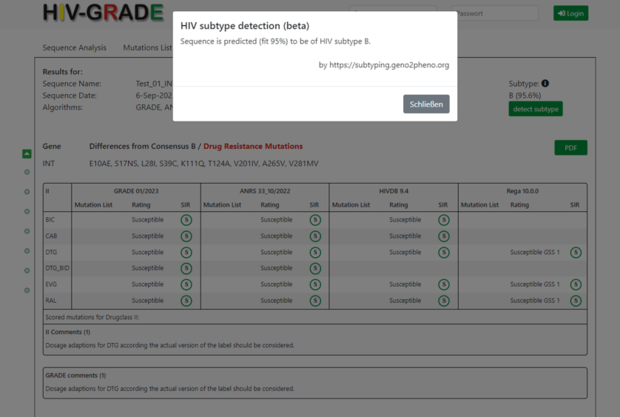
A click on the green button “detect subtype” submits the sequence automatically to the geno2pheno subtyping tool (https://subtyping.geno2pheno.org) and gives another HIV-1 subtype prediction. This is important to distinguish between HIV-1 subtypes A1 and A6, since these are not differentiated by Standford HIVdb. A click on the grey button “Schließen” closes the pop-up window and you will return to the HIV-GRADE results window.
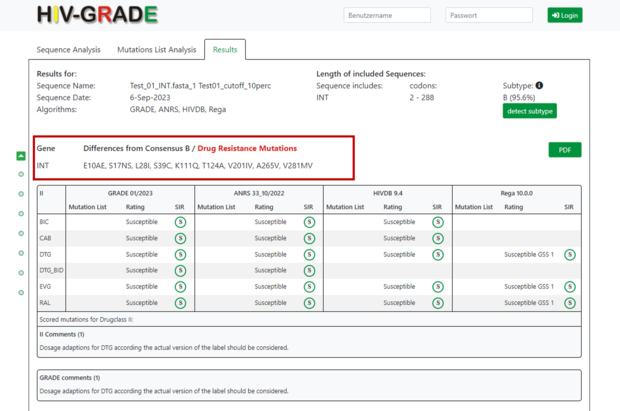
Next, you will find all amino acid differences of your submitted sequence compared to the consensus sequence of HIV-1 subtype B virus (consensus B from Los Alamos database for Protease and RT and all other genes from Stanford database). Any mutations associated with drug resistance would be highlighted in red. By moving the cursor over the mutation section, a new line will appear showing only resistance associated mutations.
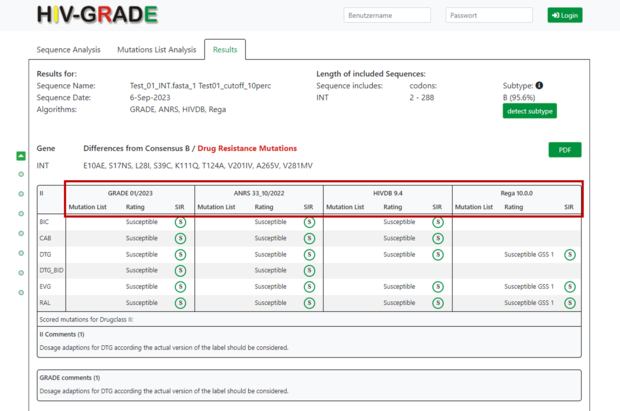
In the middle section you will find a table with result from each algorithm used for the resistance interpretation of the different antiretroviral drugs. The rows correspond to the different drugs (e.g. BIC (Bictegravir), CAB (Cabotegravir), DTG (Dolutegravir)) per drug class (NRTIs, NNRTIs, PIs, Integrase inhibitors IIs, entry inhibitors EI and/or capsid inhibitors CI) and the columns represent the result per algorithm, comprising mutation list, rating and SIR interpretation. The HIV-GRADE representation displays "intermediate resistance" instead of "possible resistance" used in the original ARNS rules. In addition, the classification “potential low-level resistance” from the original HIVdb interpretation is shown as “susceptible” in the HIV-GRADE representation, and the “low-level resistance” is summarized with the “intermediate resistance” to “intermediate resistance”.
By clicking on the comment line below each drug class section (if indicated), you will find further information on mutations and special features of algorithms.
In our example four different algorithms have been used to analyze the submitted sequence (GRADE 01/2023, ANRS 33 10_/2022, HIVDB 9.4 and Rega 10.0.0). The numbers stand for the last update or the version number.
At the bottom line of the page you will be able to archive the result or import them into LIS by downloading the results of the HIV-GRADE algorithm and alignments either as csv- or xls or pdf-file.

